Its time for a new Finestructure analysis. The difference from the earlier Finestructure analysis are more project participants and more reference populations.
CHUNKCOUNTS:
Lets first look at the chunkcounts matrix, that is the number of shared segments including both identical and related segments that have mutated (closest neighbouring segment). In the matrix the rows are receiptants and columns are the donors.
Two main branches Saami-Finns and Continental-European-Scandinavian
We see the formation of two main branches: 1) Common Continental European and Scandinavian branch 2) Saami-Finnish branch. The former branch divide again into two subbranches a Western European branch and a Eastern European branch. The Scandinavians appears to divide at the earliest branch from the Western European branch. Early branching indicate more distant relateness, late branching indicates closer relatedness. In the eastern European branch the Chuvash break away the earliest from the rest and it make sense as they are a distant population. In the latter Saami-Finnish branch the Bothnian group appears to divide the earliest from the Saami and Finns and it make sense as they likely have more Scandinavian like influence. The arrangment of trees vs other trees is arbitrary so you need to look at the heatmap and branch level to make any judgment about relationship.
(ChunkCount aggregated for individuals)
(ChunkCount aggregated with population labels)
Bothnians - two groups with different influences
In the heatmap above we see the Bothnian2 group who shows strong relationship to Scandinavains have higher sharing with the Saami. Finns do not show a similar affiliation to this group. The group share especially with the Saami2 group that have that is known to have at least partly South-Saami ancestry. Several individuals in this group also have documented Saami ancestry from the South-Saami areas. This suggest that the reason for the Bothnian2 groups seperation from other Scandinavians is due to South-Saami influence. We also see this in the PCA plot below where the Bothnian2 group close to the Scandinavians appears to pull straight towards the Saami and not to Finns.
The Bothnian1 group appears to have a somewhat different history than the Bothnian2 group, while the latter appears to be Scandinavains with Saami admixture the former appears to be more like Finns with Scandinavian admixture. We see this both in the heatmaps and in the PCA plots where there is a pull toward the Scandinavians. There also appears like this group in general appears closer to Continental-European populations than the Finns.
Finns - two groups with more western and more eastern influences.
Internally Finns divide into two groups East-Finns Finn1 and West-Finns Finn2. Eastern Finns appears to have an increased sharing between each others compared to West-Finns this probably to a late expansion into eastern and northern Finland. Finns appears to affiliate to in decreasing order Bothnian1, Bothnian2, Estonians, Scandinavians and Vologda Russians. These are all geographically close or even overlapping groups. In the finer grades we observe that Eastern Finns appears to affiliate less with Scandinavians then Western Finns. East-Finns also appears affiliate less with more western continental populations than western Finns. Western Finns seem also to be a little closer to Lithuanians than western Finns but the difference is small.
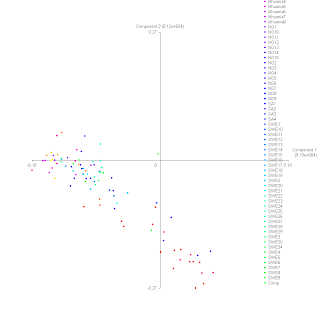
We see from the PCA plot below that Eastern Finns and Saami dominate the far right of the first dimension (horizontal) but all Finns are above the horizontal line in the second dimension (vertical). This may indicate a common genetic backgound for Saami and East-Finn in that dimension. We do not know the "original" position for East-Finns but if they originally where closer to the Saamis position on the vertical their higher position on the vertical may be due to Baltic influences, however we also see that East-Finns have the same vertical height as the Vologda Russians so it is possible that there is a direct connection between Vologda Russians and East-Finns f.ex that the Vologda Russians originally had a larger Finnsih like component that later become more similar to slavs due to centuries of migration and assimilation moving Vologda Russians to its current position.
We see further that Western Finns appears right in line between Eastern Finns and Scandinavians. This indicate that Western Finns have more Scandinavians influence also seen in the heatmap and it is the reason why Western Finns appears closer to Scandinavians. This admixture may be one factor why West-Finns appears more diverse.
We also see in the PCA plot that Estonians, Mordovians and especially the Vologda Russians appears to pull away from the Lithuanians, Belorussians and Ukrainians towards the Finns. This indicate that even these populations are continental they have a significat Finnish like components.
(ChunkCount aggregated PCA with population labels)
Saami - various degree of Scandinavian and Chuvash like influence?
The Saami divide into two groups. The Saami1 group consists of Saami entirely of North-Saami origins while the Saami2 group also contain at least partly individuals of South-Saami origin. The heat map suggest a slightly closer connection to East-Finns than to West-Finns. The Saami2 groups shows increased relationship to the Bothnian2 group than the Saami1 group. The Saami1 group shows increased affinity to the Vologda Russians and Chuvash. Both groups show same affinity to the Estonians. The Saami2 group also shows increased affinity to the Scandinavians. To sum it Saami appears to seems to affiliate with more eastern population and strongest to its closest neighbours except for the Vologda Russians and Chuvash.

The closer position of Saami2 in the PCA plot on the direct line to the Scandinavian may suggest Scandinavian influence however we do not know why East-Finns and the North-Saami group cluster to the far right. It may be the result of foundereffect and/or drift as no ancestral population can be inferred from this axis alone except it is shared by East-Finns.
On the vertical axis on the other hand we see that the Saami is at the same level as Italian, French, British and lower part of the Scandinavian cluster. It may indicate that Saami have ancestral relationship to these populations, however the positioning of the Chuvash and the increased chunk sharing indicate that the Saami maybe have been influenced vertically by a population with elements similar to the Chuvash or Vologda Russians. It may be a reflection of common Siberian like ancestry deteced in earlier analysis or an artificat of the PCA software as the Chuvash appears quite distant to the Saami in other dimensions. Further in earlier analysis there appear to be a strong correlation between Koryak, Chuvash and Vologda Russians in the Fennoscandia dataset.
Scandinavians - western, eastern and northern influences
The Scandinavians all fall into one group. Its part of the western/central european branch where it divide from the other subgroups at the earliest branch. In the heatmaps we see they are closest to the Botnian2 group that divide into a subgroup because of minority Saami ancestry. Next they affiliate to a subgroup of Brits. Then they also cluster to the Bothnian1 group that appears to be a group close to the Western Finn group pulling towards the Scandinavian cluster. Of the more distant populations the Scandinavians then cluster to the French. Then also to Finns especially the Western Finns. Scandinavians also shows increased affinity to Mordovians, Belorussians, Lithuanians and Ukrainians, actually more than Finns and Saami in general. However they have lower affinity to the Vologda Russians and Chuvash than Finns and Saami.
In the PCA plot we see that Scandinavians pull slightly away from the continental clusters towards partly the Finns and partly the Saami where they are much in line with the Belorussian-Ukrainian and the Lithuanian cluster. One may here suggest that the reason for this positioning is that Lithuanians and the Belorussian-Ukrainians do have a common backgroun in dimension but a check of the heatmap suggest that the highest intensity is between the British-French and the Finns-Saami with the former having the highest intensity. So the pull of Scandinavians from the French and the British seems to be the result of Finns and Saami influence, on the other hand one cant exclude that French and British have been pulled away from the Scandinavisn either by influenes not seen in this population panel.
On the vertical we see that the Scandinavians appears to be slighty moved upwards compared to the French and the British. This is possibly due to the pull from the Lithuanians and Belorussian-Ukrainians as Scandinavians appears to show a stronger pull to these populations in the heatmaps than to Romanians and Hungarians. If we look at the individual PCA plot we can also see that Swedes appears to dominate the upper right part while Norwegians appears to dominate the lower left part. This reflect that Swedes appears to have more eastern influences both in the direction of Lithuanians and Belorussians-Ukrainians and in the direction of Finns. The Norwegian on the other hand tend to move towards the western populations like French and the British suggesting they have more western like influences.
OTHER PLOTS AND HEATMAPS:
(ChunkCount aggregated PCA D1-D2 with individual labels)
(ChunkCount aggregated PCA D1-D3 with individual labels)
(ChunkLenght aggregated with population labels)
(ChunkLenght aggregated for individuals)

(ChunkLenght aggregated PCA D1-D2 with population labels)
(Unable to seperate the coloring for one Saami group)
(SegmentSize CL/CC aggregated for individals)
(SegmentSize CL/CC aggregated with population labels)
(SegmentSize CL/CC aggregated PCA with population labels)
(SegmentSize CL/CC aggregated PCA with individual labels)
(Updated 13 Aug 2012)