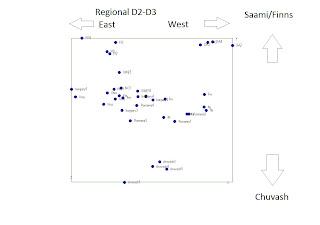
In earlier posts I have indicated that dimension 1 in the later MDS plots was of Sibieran origin. This I assumed based on similarities with the clustering with or without Siberians. I have now done a new review with the Siberian and conclude thats not the case. This is the plot in question I have published earlier. In the plot below dimension 1 (horisontal) I suggested to be a Siberian (left) vs European (right) dimension while dimension 2 I suggested to be an western European (bottom) and eastern European dimension (top). After new insights this may need some more modifications:
I did a new run with the current panel. I added 4 unrelated Sibierans (Ngaans and Dolgan) into the panel. This gave the following MDS plot:
Impossible to see the details here but the general impression is that Siberians is a fairly distant population. Dimension 1 here is the horisontal axis where the extreme left is Siberian while the extreme right is European. Dimension 2 appears to be an internal European dimension with the North Italians at one extreme while the The Finns appear on the other extreme suggesting the dimension is a north-south dimension. The Saamis appears to stick out somewhat from the Finns. In this image its hard to see the details so lets us zoom in.
As we can see here in detail the Italians have the lowest Siberian influence. It appears to be in a continuity all the way downward and increase more to left as we go down the vertical axis. The Scandinavians is inside partly in the Belorussian and Hungarian cluster. However from this part the Siberian influence separate into the Vologda Russians and the Lithuanians. Most Finns (with some internal variations) appears to have similar Siberian influence as the Vologda Russians. The Saami appears to continue further left at the same downward level as part of the Finns.
However as the plot shows above the European dimension (D2) isnt more informative than showing an North-South influence. We have an additional dimension 3 and when we compare it do dimension 2 we get a very familiar MDS plot to the one seen at the top just rotated left 90 degrees:
So basically what the earlier plot showed was that dimension was a north-south axis or more preciously a south-west vs north-east genetic and geographic distribution while dimension 2 again separate it into an west and east genetic and geographic distribution or to be more precise a north-west vs south-east genetic and geographic distribution. Below dimension 1 represented geographically.
Dimension SNP (and variation):
D1: 51 706 SNP (10,6%)
D2: 17 341 SNP (3,6%)
D3: 9 758 SNP (2,0%)
All grandparents born in Norway, Sweden or Finland? Email your 23andme/FamilyFinder/deCODEme genotype file zipped to tjaaehkere at yahoo.no. NEW! Estonians, Lithuanians, Latvians, Germans, Nederlands, Belgians, Luxenbourg, Austrians, Checkz, Slovaks, Balkans, Danes, Poles, Icelanders and Russians also welcome! ANALYSIS STATUS: FILE PREPERATIONS!
Totalt antall sidevisninger
torsdag 1. desember 2011
tirsdag 29. november 2011
Geographic MDS maps of Fennoscandia and Europe (updated)
There exists some similarity between Finns and Lithuanians not seen in Saamis, so when adding the Lithuanians the Finns moved out of the North-Saami cluster compared to the previous analysis. Therefore Lithuanians where added. Also FI5 and FI6 where 2nd cousins pulled down the Finns into the North-Saami cluster. Removing each indepentently resulted in the same effect suggesting the relatedness had an effect in the analysis. FI6 was therefore removed.
The MDS plots can be confusing for some. I therefore made a map showing the geographic average distributions for the 3 main components and it may show an intriging history. Dimension 1 accounts for the most of the variation, while dimension 2 account for the second largest genetic variantion while dimension 3 account for the third largest genetic variation. Therefore the dimensions do not acount equally when represented in a 3D view.
The map appears to show Saamis as extremes in all three dimensions.
This is how much each dimension explains the genetic variation in each dimension:
D1: 20 604 SNP 4.2%
D2: 10 417 SNP 2.1%
D3: 8 560 SNP 1.8%
Dimension Total: 8.1%
This is how much each dimension explains the genetic variation in each dimension:
D1: 20 604 SNP 4.2%
D2: 10 417 SNP 2.1%
D3: 8 560 SNP 1.8%
Dimension Total: 8.1%
D1: Saamis, Finns and Vologda Russians appears to make a far eastern bridge. Earlier analysis seem to suggest that this is a Siberian component seen in Europe. It appears absolutely lowest among the Italians. Note that the Italians appears to be "isolated" in Europe in this context with much higher rates seen in close populations. Scandinavians appear closest to Belorussians and Hungarians in this dimension.
D2 also appears very high among the Saami and seem to have a western European distribution as it has high frequency also among Finns, Swedes and Norwegians. It is also higher in western Europe as seen among the French and Italians. However it drops very dramatically from Finns to Lithuanians, Estonia and Vologda Russian. The level increases again in more western east-European populations like Hungarians and Romanians.
D3 also appears very high among the Saami but shows an intriguing pattern. Its very low among Scandinavians, Lithuanians and Estonians, however the next highest occurrences are found among the Italians, then the Romanians and the Vologda Russians and then finally Finns just somewhat above intermediate levels like the French, Hungarians and Belorussians.
May it be the "eastern component" researcher Tambets mentions in her abstract for the coming paper about genetic of the uralic speakers? If yes why is it also found second highest among Italians?
Also the local opposite clustering of this component only seen among Norwegians, Swedes, Lithuanians and Estonians are intriguing suggesting something in common.
As the result indicate the Saami outlierness appears to be the sum of these 3 different dimensions or genetic components. Italians appears to be opposite outlier in D1, Lithuanians appears to be opposite outlier in D2 and Swedes appears to be opposite outlier in D3.
fredag 25. november 2011
Fennoscandia BGA regional MDS update (2nd update)
We got 5 new project members since last update. SWE17, SWE18, SWE19. FI12 and FI13. The interpretations are much the same as given in earlier posts. We can see that SWE14 and SWE17 are outside the main Scandinavian cluster pulling toward the eastern European populations. SWE19 appears to cluster well into the Scandinavain cluster. Interesting that FI12 a southern Finn appears close to the Estonian ES1 halfway towards the Vologda Russians and the Belorussians. FI13 have ancestry from Savo and cluster in the Karelian cluster.
We also see some slight movements of others after adding more individuals. This is normal as adding more individuals influence the positioning of all other individuals especially the closer ones to new added individuals.
tirsdag 22. november 2011
Fennoscandian Mutation Sharing Matrix
All participants have received the Mutation Sharing Matrix of currently known unique haplotype mutations map for Fennoscandia shared with two or more individuals. These mutations is currently only known in Fennoscandia.
Its highly recommended to use Excel 2007 or later when using. Enclosed Excel 2007 and Excel 2003 versions.
X-axis - Individuals
Y-axis - Clusters
How to use:
1. Find your code at the top.
2. Hit the filter and choose "1".
3. You now have a overview over at what cluster and to what other individuals you share mutations with.
4. To the far right is a indicative distribution of the cluster etnical distribution. Please use with extreme caution in regards to interpretation.
Other things:
* Individuals of known partly Saami ancestry is indicated with blue colour.
* Individuals of known partly Saami ancestry is labeled as Norwegians and Swedes.
* Interpretation of mutation patterns must be done with extreme caution. There are many possible problems like etnical categorisations for haplotypes that is also shared outside ethical boundaries but within geographical boundaries. The mutation could be "native" but also arrived trough later migration.
* Widespread distributions may indicate higher age for the mutation cluster or it may indicate a common immigrant who hit the genetic jackpot.
If you have not received the sheets please contact me on email.
Anders
Its highly recommended to use Excel 2007 or later when using. Enclosed Excel 2007 and Excel 2003 versions.
X-axis - Individuals
Y-axis - Clusters
How to use:
1. Find your code at the top.
2. Hit the filter and choose "1".
3. You now have a overview over at what cluster and to what other individuals you share mutations with.
4. To the far right is a indicative distribution of the cluster etnical distribution. Please use with extreme caution in regards to interpretation.
Other things:
* Individuals of known partly Saami ancestry is indicated with blue colour.
* Individuals of known partly Saami ancestry is labeled as Norwegians and Swedes.
* Interpretation of mutation patterns must be done with extreme caution. There are many possible problems like etnical categorisations for haplotypes that is also shared outside ethical boundaries but within geographical boundaries. The mutation could be "native" but also arrived trough later migration.
* Widespread distributions may indicate higher age for the mutation cluster or it may indicate a common immigrant who hit the genetic jackpot.
If you have not received the sheets please contact me on email.
Anders
mandag 21. november 2011
Accurate method for pinpointing autosomal ancestry?
In the earlier post I reported to have found 2 haplotype clusters seen in 6 individuals each. I here look closer at 1 of those 2 clusters to see what can be inferred from them.
The core haplotype
The haplotype identified occured in 6 individuals: 3 Finns, 2 Swedes and 1 Norwegian (no Saamis) and no one else in the population panel. The core of the haplotype is GC with the markers rs6683734 and rs4649296. These are together mutations that can only be found among Fennoscandians. However the spread is very limited only occuring in 6 of 88 haplotypes (or "3" of 44 individuals).
The extented haplotype
The haplotype can be furter extended for all with these markers: rs16858853 rs12087818 rs701177 rs16858884 rs701176 rs701173 rs11800619 rs10489806 rs7546115 rs10910120 rs1033322 rs12124323 rs17752790 rs6697791 rs12140862 rs12130755 rs6683734 rs4649296. The physical distance between the first and last is 75kb or 0.075 Mb. The individual haplotype appears to have gone trough recombination if extending the haplotype further.
The extended haplotypes founder haplotype appears to have accumulated several mutations that appears to have some local variation suggest it has a age and history in Fennoscandia seen in total. Two Finns FI11 and FI12 appears to share what appears to be the founder haplotype. FI10 have accumulated two mutations not seen in anywhere else. The SWE1 and SWE13 haplotypes appears to have separate history from the founderhaplotype seen in the two Finns. Here the SWE1 appears to be the founder while the SWE13 appears as a subfounder. The one Norwegian haplotype NO5 appears to have a history closest to the Finn haplotype.
The mutation diversity suggest a history even its widespread. The founder haplotype is seen in Finland while 4 of 6 mutations is seen in Swedes and a Norwegian. The Swedes even have a sub-node haplotype. This suggest that Sweden is the oldest location for this haplotype with Finland as runner up. It may be youngest in Norway but belong to the Scandinavian cluster in other analysis so it may be grouped with Swedes. More samples could possibly enlight the question of origin.
The ancestral haplotype
It would be of great interest to identify the true ancestral haplotype to the above described haplotype in Continental-Europe to be able to infer the "point" of entry to Fennoscandia.
I managed to extract this possible ancestral haplotypes. Two from Spain and 1 from the French Basque. As seen from these it appears like third last and last SNP mutated in Fennoscandia from AGA to GGC. The Norwegian's AGC may be due to back mutations, error or it could be transition between the third last and and last SNP. If correct this could mean the point of entry was Norway but it is inconsistent with extended haplotype having the lowest diversity but not if we group Swedes and Norwegians.
The SNP before the last tree is the common founder haplotype. I attempted to find sign of diversity for the Iberians for this part but didnt manage to detect any at first screening. The zero diversity of the Iberian group for this part compared to the Fennoscandian suggest the Fennoscandian haplotype are older. The difference is mutated positions for the third last and last SNP.
SUMMARY: As seen from this posting trying to reconstruct a autosomal haplotype could be complex but give accurate and informative genetic history. The core haplotype appears to be simple and effective in proving Fennoscandinavian ancestry despite its tiny size, while the extended haplotype can provide clues to its age and history.
This method using core haplotypes with limited distributed mutations appears to be very accurate in pinpointing ancestry
The core haplotype
The haplotype identified occured in 6 individuals: 3 Finns, 2 Swedes and 1 Norwegian (no Saamis) and no one else in the population panel. The core of the haplotype is GC with the markers rs6683734 and rs4649296. These are together mutations that can only be found among Fennoscandians. However the spread is very limited only occuring in 6 of 88 haplotypes (or "3" of 44 individuals).
The extented haplotype
The haplotype can be furter extended for all with these markers: rs16858853 rs12087818 rs701177 rs16858884 rs701176 rs701173 rs11800619 rs10489806 rs7546115 rs10910120 rs1033322 rs12124323 rs17752790 rs6697791 rs12140862 rs12130755 rs6683734 rs4649296. The physical distance between the first and last is 75kb or 0.075 Mb. The individual haplotype appears to have gone trough recombination if extending the haplotype further.
The extended haplotypes founder haplotype appears to have accumulated several mutations that appears to have some local variation suggest it has a age and history in Fennoscandia seen in total. Two Finns FI11 and FI12 appears to share what appears to be the founder haplotype. FI10 have accumulated two mutations not seen in anywhere else. The SWE1 and SWE13 haplotypes appears to have separate history from the founderhaplotype seen in the two Finns. Here the SWE1 appears to be the founder while the SWE13 appears as a subfounder. The one Norwegian haplotype NO5 appears to have a history closest to the Finn haplotype.
The mutation diversity suggest a history even its widespread. The founder haplotype is seen in Finland while 4 of 6 mutations is seen in Swedes and a Norwegian. The Swedes even have a sub-node haplotype. This suggest that Sweden is the oldest location for this haplotype with Finland as runner up. It may be youngest in Norway but belong to the Scandinavian cluster in other analysis so it may be grouped with Swedes. More samples could possibly enlight the question of origin.
The ancestral haplotype
It would be of great interest to identify the true ancestral haplotype to the above described haplotype in Continental-Europe to be able to infer the "point" of entry to Fennoscandia.
I managed to extract this possible ancestral haplotypes. Two from Spain and 1 from the French Basque. As seen from these it appears like third last and last SNP mutated in Fennoscandia from AGA to GGC. The Norwegian's AGC may be due to back mutations, error or it could be transition between the third last and and last SNP. If correct this could mean the point of entry was Norway but it is inconsistent with extended haplotype having the lowest diversity but not if we group Swedes and Norwegians.
The SNP before the last tree is the common founder haplotype. I attempted to find sign of diversity for the Iberians for this part but didnt manage to detect any at first screening. The zero diversity of the Iberian group for this part compared to the Fennoscandian suggest the Fennoscandian haplotype are older. The difference is mutated positions for the third last and last SNP.
SUMMARY: As seen from this posting trying to reconstruct a autosomal haplotype could be complex but give accurate and informative genetic history. The core haplotype appears to be simple and effective in proving Fennoscandinavian ancestry despite its tiny size, while the extended haplotype can provide clues to its age and history.
This method using core haplotypes with limited distributed mutations appears to be very accurate in pinpointing ancestry
fredag 18. november 2011
Autosomal Haplotype Clustering Patterns - Actual or Error? (updated)
I have found this pattern of haplotype clustering within individuals from Norway, Sweden and Finland on Chr 1 using 38.5k SNP:
Unique haplotypes not shared with others - 1 344 (typical between 15 to 50 per individual)
Haplotypes shared between 2 ind - 157
Haplotypes shared between 3 ind - 30
Haplotypes shared between 4 ind - 3
Haplotypes shared between 5 ind - 2
Haplotypes shared between 6 ind - 2
As this shows widespread haplotype clusters are much rarer than those shared with only two individuals, but the "unique" haplotype clusters appears to be absolutely highest at the individual level.
This raises the questions why its like this. I suspect its the following reasons:
1. The effect of recombination splitting or killing haplotypes. However the maximum haplotype size in clusters is 500 SNP. Redusing it to 100 SNP only reduced to 1324 unique haplotype clusters. Reducing to max 10 SNP only reduced to 1189 unique haplotype clusters. Reducing to 5 SNP reduced only to 962 unique haplotype clusters. If reducing to 2 SNP only 277 unique haplotype clusters.
2. The effect of limited population data. Its possible more individuals and populations would reduce the number of unique haplotype clusters.
3. The effect of undetected errors in the genotypes. However no correlation between high unique haplotypes found in individuals and high detected genotype error rate for these.
4. The effect of incorrect phasing as the result of errors in genotype or/and ordinary phasing error as result of the model used.
5. The effect of haplotype or mutation extinction. Recent individual haplotypes or mutations have limited spread generally, while older haplotype clusters or mutations have larger geographic spread.
So what I infer from this is that these unique haplotype clusters is rather small and not very large. These numbers have been generated from software made for finding genetic diseases from haplotypes where you mark individuals with certain traits cases and check them vs the controls. If there is any haplotype strongly associated with a trait the associated haplotype is found. These haplotypes are usually not very large. Just check the SNP used by 23andme health section.. So is also the cases with these haplotypes.
The software do for many haplotypes infer parent-child relationships between them indicating that haplotype mutations are in the picture at least when I check at the individual level.
Unique haplotypes not shared with others - 1 344 (typical between 15 to 50 per individual)
Haplotypes shared between 2 ind - 157
Haplotypes shared between 3 ind - 30
Haplotypes shared between 4 ind - 3
Haplotypes shared between 5 ind - 2
Haplotypes shared between 6 ind - 2
As this shows widespread haplotype clusters are much rarer than those shared with only two individuals, but the "unique" haplotype clusters appears to be absolutely highest at the individual level.
This raises the questions why its like this. I suspect its the following reasons:
1. The effect of recombination splitting or killing haplotypes. However the maximum haplotype size in clusters is 500 SNP. Redusing it to 100 SNP only reduced to 1324 unique haplotype clusters. Reducing to max 10 SNP only reduced to 1189 unique haplotype clusters. Reducing to 5 SNP reduced only to 962 unique haplotype clusters. If reducing to 2 SNP only 277 unique haplotype clusters.
2. The effect of limited population data. Its possible more individuals and populations would reduce the number of unique haplotype clusters.
3. The effect of undetected errors in the genotypes. However no correlation between high unique haplotypes found in individuals and high detected genotype error rate for these.
4. The effect of incorrect phasing as the result of errors in genotype or/and ordinary phasing error as result of the model used.
5. The effect of haplotype or mutation extinction. Recent individual haplotypes or mutations have limited spread generally, while older haplotype clusters or mutations have larger geographic spread.
So what I infer from this is that these unique haplotype clusters is rather small and not very large. These numbers have been generated from software made for finding genetic diseases from haplotypes where you mark individuals with certain traits cases and check them vs the controls. If there is any haplotype strongly associated with a trait the associated haplotype is found. These haplotypes are usually not very large. Just check the SNP used by 23andme health section.. So is also the cases with these haplotypes.
The software do for many haplotypes infer parent-child relationships between them indicating that haplotype mutations are in the picture at least when I check at the individual level.
mandag 7. november 2011
How genetic similar or disimar where your parents ancestry? (updated)
How genetic similar or dissimilar your mother and father ancestry is of some interest in infering your past genetic history. In scientific language its called "Runs of homozygosity" or simply "ROH" and are segments of identical blocks of DNA on your autosomes that you received from both of your parents.
In one hypothetical extreme if your parents where closely related like cousins you would have a few large segments of ROH or if both came from the same population isolate or village but not recently related, many small ROH segments. On the other side if your parents where of very different origin you would probably have very few and short ROH segments. In fact ROH works similar to 23andme's Relative Finder by finding common segments between your parents. If sharing many segments, probably related, if sharing few or no segment not related.
ROH therefore provides insight on whether your ancestors derived from a small isolated population, were of mixed or urban origins, or even if there was consanguinity in your lineage.
I have attempted to reconstruct a similar analysis provided by Etnoancestry using PLINK's ROH functionality. I used the following parameters (default settings):
Min ROH size: 1 Mb
Min ROH SNP: 100
Min ROH Density: Kb/SNP: 50
Largest ROH Gap: 1 Mb
Number of SNP: 530k, 22 autosomes, no pruning.
The ROH run for each participant is presented in the graph below. As shown there appears to be great variation between the samples. The Y-axis represent the number of ROH segments found while the X-axis represent the sum size in Mb for all those found ROH's. In the one extreme FI10 and SWE16 is found to have the largest number of ROH's and the largest total size of ROH or in other words they had parents with the most similar ancestry. On the other extreme you find NO7 and NO11 both of mixed backgrounds, the first Norwegian-Saami mix and the second Norwegian-Swedish mix (and possibly some minor Saami/Finnic mix), in other words they had the parents with the most dissimilar ancestry.
So then the question naturally of course arise, do these individuals with high ROH is the result of recent inbreeding or consanguinity? The answer can be infered by calculating the average segment size by dividing the total size of ROH with the number of ROH found. If the calculation result in a few large segments its indication of recent consanguinity as few recombination events have divided the ROH into smaller pieces since the consanguinity, however if the calculation result in many number of small ROH its an indication that your came from a small population isolate with little consanguinity in recent times as recombination have split the ROH segments into many smaller pieces.
The calculation of the average ROH block size among all members appears consistent with a scenario with no recent consanguinity for all participants (visually confirmed with actual data). To illustrate it SA2 who score very high in both number and sum of ROH but the average ROH size isnt so much different from NO7 who's have the least similar ROH.
In MDS plots having a individual with extreme ROH would have similar effects like having related samples resulting in own clustering. In 23andme's Relative Finder individuals of from same population isolate would appear artificially as closer relatives than they actually where.
In one hypothetical extreme if your parents where closely related like cousins you would have a few large segments of ROH or if both came from the same population isolate or village but not recently related, many small ROH segments. On the other side if your parents where of very different origin you would probably have very few and short ROH segments. In fact ROH works similar to 23andme's Relative Finder by finding common segments between your parents. If sharing many segments, probably related, if sharing few or no segment not related.
ROH therefore provides insight on whether your ancestors derived from a small isolated population, were of mixed or urban origins, or even if there was consanguinity in your lineage.
I have attempted to reconstruct a similar analysis provided by Etnoancestry using PLINK's ROH functionality. I used the following parameters (default settings):
Min ROH size: 1 Mb
Min ROH SNP: 100
Min ROH Density: Kb/SNP: 50
Largest ROH Gap: 1 Mb
Number of SNP: 530k, 22 autosomes, no pruning.
The ROH run for each participant is presented in the graph below. As shown there appears to be great variation between the samples. The Y-axis represent the number of ROH segments found while the X-axis represent the sum size in Mb for all those found ROH's. In the one extreme FI10 and SWE16 is found to have the largest number of ROH's and the largest total size of ROH or in other words they had parents with the most similar ancestry. On the other extreme you find NO7 and NO11 both of mixed backgrounds, the first Norwegian-Saami mix and the second Norwegian-Swedish mix (and possibly some minor Saami/Finnic mix), in other words they had the parents with the most dissimilar ancestry.
So then the question naturally of course arise, do these individuals with high ROH is the result of recent inbreeding or consanguinity? The answer can be infered by calculating the average segment size by dividing the total size of ROH with the number of ROH found. If the calculation result in a few large segments its indication of recent consanguinity as few recombination events have divided the ROH into smaller pieces since the consanguinity, however if the calculation result in many number of small ROH its an indication that your came from a small population isolate with little consanguinity in recent times as recombination have split the ROH segments into many smaller pieces.
The calculation of the average ROH block size among all members appears consistent with a scenario with no recent consanguinity for all participants (visually confirmed with actual data). To illustrate it SA2 who score very high in both number and sum of ROH but the average ROH size isnt so much different from NO7 who's have the least similar ROH.
In MDS plots having a individual with extreme ROH would have similar effects like having related samples resulting in own clustering. In 23andme's Relative Finder individuals of from same population isolate would appear artificially as closer relatives than they actually where.
fredag 4. november 2011
Investigating the genetic background of Finns
We continue the analysis of the latest MDS run now looking at the Finns. In the plot it appears that Finns divide into several groups with different origins.
The first group is the Karelian group. These are individuals with most of their background from from Finnish Karelia. FI10 is entirely North-Karelian. This cluster appear to have somewhat lower Siberian influence than the Saami but with more similarites with Vologda Russians than other Finns as they appear to pull in the direction from the North-Saami and below mentioned Finns. FI2 who is outside this cluster pull even more towards the Vologda Russian cluster.
The second group is a more mixed group with ancestry from Tornea river area, Oulu, Lappi, Finnsih Karelia, Central-Bothna, NW, SW and Central Finland. FI5 who have ancestry from Western Finland pull slightly toward the Scandinavian cluster. The proximity of this cluster to the North-Saami in D2 is interesting it could mean Saamis have a close common background with this cluster in this dimension. However in D3 (not shown) this affiliation disappear. Else these Finns appear to pull slightly towards both Scandianvians (especially FI5) while FI3 and FI6 pull towards the Karelians and The Vologda Russians. Interestingly in linguistic research the Saami self designation is suggested to be come from the Häme (Tavastia) region in West/Central-Finland.
The third group is F4 and FI9 who appear together to form a Bothnian cluster. Both have most or all of their background from this area. This group appear to be intermediate between the second Finnish group and the Scandinavian cluster. NO6 position here is the result of this individual beeing a mixture of Scandinavian and North-Saami and so is only artificially in this cluster, as in D3 this affiliation vanish while FI4 and FI9 stays put to each other in D3 (not shown) between the second Finnish group and Scandinavian. FI9 appears to pull more towards the Swedes while FI4 appears to point towards the "other end" of the Scandinavian cluster.
The fourth group is F1 who is the only known Swedish Finn in this project. This individual is between the Scandinavian and the Bothian Finnish group and appears to point further at the second Finnish group. In D3 this individual appears to pull strongly towards the Swedes in the Scandinavian cluster.
SUMMARY: So to conclude it appears that Finns have both common and different origins. Finns appear in these dimensions to divide into two groups. All Finns appears to have something in common with the Saami in D2 however western Finns and Eastern Finns have different external influences. Western Finns appear to be more influenced by Scandinavians while eastern Finns appears more influenced by the Vologda Russians.
The first group is the Karelian group. These are individuals with most of their background from from Finnish Karelia. FI10 is entirely North-Karelian. This cluster appear to have somewhat lower Siberian influence than the Saami but with more similarites with Vologda Russians than other Finns as they appear to pull in the direction from the North-Saami and below mentioned Finns. FI2 who is outside this cluster pull even more towards the Vologda Russian cluster.
The second group is a more mixed group with ancestry from Tornea river area, Oulu, Lappi, Finnsih Karelia, Central-Bothna, NW, SW and Central Finland. FI5 who have ancestry from Western Finland pull slightly toward the Scandinavian cluster. The proximity of this cluster to the North-Saami in D2 is interesting it could mean Saamis have a close common background with this cluster in this dimension. However in D3 (not shown) this affiliation disappear. Else these Finns appear to pull slightly towards both Scandianvians (especially FI5) while FI3 and FI6 pull towards the Karelians and The Vologda Russians. Interestingly in linguistic research the Saami self designation is suggested to be come from the Häme (Tavastia) region in West/Central-Finland.
The third group is F4 and FI9 who appear together to form a Bothnian cluster. Both have most or all of their background from this area. This group appear to be intermediate between the second Finnish group and the Scandinavian cluster. NO6 position here is the result of this individual beeing a mixture of Scandinavian and North-Saami and so is only artificially in this cluster, as in D3 this affiliation vanish while FI4 and FI9 stays put to each other in D3 (not shown) between the second Finnish group and Scandinavian. FI9 appears to pull more towards the Swedes while FI4 appears to point towards the "other end" of the Scandinavian cluster.
The fourth group is F1 who is the only known Swedish Finn in this project. This individual is between the Scandinavian and the Bothian Finnish group and appears to point further at the second Finnish group. In D3 this individual appears to pull strongly towards the Swedes in the Scandinavian cluster.
SUMMARY: So to conclude it appears that Finns have both common and different origins. Finns appear in these dimensions to divide into two groups. All Finns appears to have something in common with the Saami in D2 however western Finns and Eastern Finns have different external influences. Western Finns appear to be more influenced by Scandinavians while eastern Finns appears more influenced by the Vologda Russians.
torsdag 3. november 2011
Finding South-Saami ancestry in Scandinavians
It appears that the local analysis been done so far had weaknesses. It appears not managed to catch all people of at least partly South-Saami ancestry. I suspect the reason have been in the MDS program itself with different sample sizes in clusters and numbers of clusters affecting the result especially for partly mixed individuals. When including more outside populations it appears to make genetic distinctions clearer within Fennoscandia possibly from the earlier mentioned reasons.
Ref pop: French, Italians, Hungarians, Romanians, Vologda Russians, Norwegians, Swedes, Finns and Saami.
Number of SNP: 530k (not pruned) - 22 autosomal chromosomes.
PLINK MDS plot dimensions: 3
In the earlier local analysis it appeared obviously that samples NO6, NO7 and SWE7 must have been at least of partly Saami background with especially SWE7 plotting "weird", but not as obvious for sample SWE11 who earlier in the local analysis showed some weak pull out of the Scandinavian cluster. In this newest MDS run however SWE11 seperated very clearly from the Scandinavian cluster together with unrelated NO6 and SWE7.
Lets look first at the new MDS plot. In this plot top-bottom axis appears to be east and west geneticially speaking with the Vologda Russians and Belorussians at the top edge representing the most eastern population and Saamis at first glance the most "western" population with following Finns, Scandinavians and French.
Lets look at the second dimension in the left-right axis. This appears to reflect Siberian influence in Europe. The Saamis and many Finns appears to have the strongest influence from Siberia with Vologda Russians, Belorussians and Scandinavians following. The lowest Siberian influence appears to be among the Italians, French and Romanians.
So what differentiate North-Saami from South-Saami? It can be shown in the plot below. In Dimension 2 (D2) where the North-Saami and South Saami cleary is alone at the lower part of the plot in the extreme "West". Obviously both Saami group share ancestry in this common European dimension. This picture however change when we compare to Dimension 1 (D1) that shows levels of Siberian ancestry. Here the North-Saami peaks together with eastern Finns to the far right, while the South-Saami have levels comparable to Swedish Finns.
About the South Saami samples: SWE7 is confirmed at least partly of Saami background but not of North Saami background. SWE11 have also some geneological confirmed minor Saami background and have most of the origin from current and earlier known non-North-Saami areas. No information is known about NO7.
As comparisment NO6 who have geogaphical origin in the North-Saami area seperate very clearly from NO7, SWE7 and SWE11 by pulling straight at the North-Saami cluster (SA1-SA4) and appearing immediate between the Scandinavain and North-Saami cluster demonstrating that the Saami origin is different for NO7, SWE7 and SWE11.
So apparantly SWE7, SWE11 and NO7 have South-Saami background.
Ref pop: French, Italians, Hungarians, Romanians, Vologda Russians, Norwegians, Swedes, Finns and Saami.
Number of SNP: 530k (not pruned) - 22 autosomal chromosomes.
PLINK MDS plot dimensions: 3
In the earlier local analysis it appeared obviously that samples NO6, NO7 and SWE7 must have been at least of partly Saami background with especially SWE7 plotting "weird", but not as obvious for sample SWE11 who earlier in the local analysis showed some weak pull out of the Scandinavian cluster. In this newest MDS run however SWE11 seperated very clearly from the Scandinavian cluster together with unrelated NO6 and SWE7.
Lets look first at the new MDS plot. In this plot top-bottom axis appears to be east and west geneticially speaking with the Vologda Russians and Belorussians at the top edge representing the most eastern population and Saamis at first glance the most "western" population with following Finns, Scandinavians and French.
Lets look at the second dimension in the left-right axis. This appears to reflect Siberian influence in Europe. The Saamis and many Finns appears to have the strongest influence from Siberia with Vologda Russians, Belorussians and Scandinavians following. The lowest Siberian influence appears to be among the Italians, French and Romanians.
So what differentiate North-Saami from South-Saami? It can be shown in the plot below. In Dimension 2 (D2) where the North-Saami and South Saami cleary is alone at the lower part of the plot in the extreme "West". Obviously both Saami group share ancestry in this common European dimension. This picture however change when we compare to Dimension 1 (D1) that shows levels of Siberian ancestry. Here the North-Saami peaks together with eastern Finns to the far right, while the South-Saami have levels comparable to Swedish Finns.
About the South Saami samples: SWE7 is confirmed at least partly of Saami background but not of North Saami background. SWE11 have also some geneological confirmed minor Saami background and have most of the origin from current and earlier known non-North-Saami areas. No information is known about NO7.
As comparisment NO6 who have geogaphical origin in the North-Saami area seperate very clearly from NO7, SWE7 and SWE11 by pulling straight at the North-Saami cluster (SA1-SA4) and appearing immediate between the Scandinavain and North-Saami cluster demonstrating that the Saami origin is different for NO7, SWE7 and SWE11.
So apparantly SWE7, SWE11 and NO7 have South-Saami background.
onsdag 2. november 2011
Little Study of the Saami, Finns and Scandinavians
As large sample sizes have a tendency to drown the 4 sample Saami clustering I have done a small study using only 4 individuals from each population to try to infer the Saamis relationship with other populations. Only 12 of the 41 participants is included in this analysis but its interferance should apply to the rest in general depending on your clustering in the earlier posten local plots using only Norwegians, Swedes, Finns and Saami.
What have been used:
Ref pop: French, Italians, Hungarians, Romanians, Vologda Russians, Chuvash, Norwegians, Swedes, Finns and Saami. Siberians: Ngan, Dolgans. 4 samples from each cluster.
Number of SNP: 530k (not pruned) - 22 autosomal chromosomes.
PLINK MDS plot dimensions: 3
EUROPE ONLY
Dimension plot D1-2:
1. D1: This dimension on the right-left axis may reflect possible Siberian ancestry levels among the populations. Two Saamis reach the same level as the two of the Chuvashes. Finns follows close after together with two Vologda Russians. To the far left we find Italians who are most geographical distant appears to have the least of this influence. See also Europe plus Siberian plot including four unadmixted Siberians.
2. D2: This dimension on the top-bottom axis may reflect extremes between the Saami and Finns on the top and the Chuvashes at the bottom and where contintal europeans and Scandinavians appear as intermediate but closer to Saamis and Finns. Saamis and Finns are at the same level in this dimension suggesting a common background in this dimension while central-europeans share considerable more with the Chuvashes maybe from eastern influences that didnt reach the Saamis and Finns to the same extent.
Dimension plot D2-3:
3. D2: The top-bottom axis is the same as in D2 so the comment there is the same here.
4. D3: This right-left dimension appears to divide the Saami and the Finns. Here the Saami share the far right with 1 French and 2 Italians and 1 Romanians as a close follow up in what appears at first as the western or central part of a continental european cluster. The Finns appears on the left side of the continental cluster that appears as the eastern part of a continental europeal cluster together with Vologda Russians and Scandinavians. This suggest that Finns and Scandiavians have a more eastern origin than the Saami in this dimension who appears far western.
SUMMARY EUROPE ONLY:
* Saami at first glance appears to have the largest "Siberian" ancestry followed by Finns and then Vologda Russians. Scandianvians appears at the same level as central-europeans. (D1)
* Saami and Finns appears to share ancestry. Scandianvians and Continental Europeans and Scandinavians appears to have more eastern influences pulling them closer to the Chuvashes. (D2)
* Saami have what appears at first to be a western component pulling them far "west" than both Finns and Scandinavians who appears as far "east" as Vologda Russians. (D3).
-> This analysis may suggest that part of the "West" origin of the Saami do not have its root in recent Scandianvian admixture.
EUROPE PLUS SIBERIANS:
Dimension plot D1-2:
1. D1: This dimension on the left-right axis appears to be between Siberians to the far left and europeans to the far right. The Saami pull to the left the most after the Chuvash then followed equally Finns and Vologda Russians. Scandinavians appears to stay the same as continental-europeans in this respect see comment D1 in the EUROPE ONLY analysis.
2. D2: This dimension on the top-bottom axis appears to be with the Saami in the one extreme and the Italians at the bottom. Finns follows the Saami just after. The Vologda Russians and the Chuvash fills the void between the Saami and Finns vs the Scandinavians who appears to be at the "northernmost" part of the long continental european cluster. The Vologda Russian cluster appears to bridge to the Finns. Siberians appears here to have something in common with central-europeans maybe admixture however in ADMIXTURE runs the selected 4 Siberians do not appear to have european admixture.
Dimension plot D2-3:
3. D2: Same as D2 in Dimension plot D2.
4. D3: The left-right axis appears to be a east and west axis placing Chuvashes to the far left. Finns appear a little more to the left than the Saami. Scandinavians appears to pull even more to the left togheter with what appears to be central european cluster. Vologda Russians appears imidate between Chuvashes and Central-Europe.
SUMMARY EUROPE WITH SIBERIANS
* Saami at first glance appears to have the largest "Siberian" ancestry followed by Finns and then Vologda Russians. Scandianvians appears at the same level as central-europeans (D1):
* Saami appears far west in D3 while Finns and especially Scandinavians cluster with central-Europe.
* Saami appears uppermost or norternmost in the D2 plot followed by Finns, Vologda Russians/Chuvash and Scandinavians, then central-europeans and southern Europeans at the bottom.
tirsdag 7. juni 2011
Updated local Fennoscandia analysis
-- 3 new participanst: 2 from Sweden and 1 from Finland.
-- Changable 3D plot for D1-3 sendt to all participants by email.
-- Else nothing new to comment.
Have questions or want to participate? Send email to tjaaehkere at yahoo.no
tirsdag 19. april 2011
A SMALL COMPARISMENT OF LAMP, ADMIXTURE AND AIMs
INTRODUCTION
The LAMP 2.5 genetic analysis software picks AIM's in its analysis based on the allele frequences given to it. On this basis I would like to check how LAMP perform by comparing to the widely used ADMIXTURE software. Of special interest are if ADMIXTURE clusters individuals similar to LAMP.
METHOD
I first estimatet the ancestral frequency data for Norwegians, Swedes, Saami and Finns from a subset of individuals clustering to these based on their MDS plots. I then run LAMP on recommended settings except this time I run it assuming the populations had been mixing for 10 generations the same presumption used in ADMIXTURE. I then picked the AIM's chosen for this analysis by LAMP and used them in a run for the whole dataset of 36 individuals.
OBSERVATIONS:
* All the Finns who clustered close to or 100% in LAMP also did the same in ADMIXTURE.
* In LAMP 9 Norwegians appeared 100% or close to 100% Norwegian. In ADMIXTURE 2 of these 9 appeared strongly mixed with Saami and Finns in ADMIXTURE.
* Saami all appear in LAMP close to or at 100% Saami. In ADMIXTURE only 1 appears as heavily mixed.
* All the Swedes who appeared as 100% Swedish in LAMP was also classified as 100% Swedish in ADMIXTURE.
* The Norwegian mixture in Swedes and Finns appears stronger in LAMP than in ADMIXTURE for those individuals who have it.
* Saami amix proportions seen in considerabe proportions in ADMIXTURE are often very little or gone in LAMP.
COMMENT:
It seems LAMP and ADMIXTURE is largely consistent about the "pure breed" and supports that the ancestral allele frequency data used in LAMP are correct. What LAMP and ADMIXTURE do disagree some about are the admixtures. 3 individuals who in ADMIXTURE was classified as mixed was not or very little admixted in LAMP. The reason for this are that LAMP and ADMIXTURE uses different methods to estimate admixture.
ADMIXTURE basically estimate allele freqency differences between SNP that is assumed to be independent and than cluster it. It does not care about physical positions of SNP, cM (centimorgans) between SNP and recombination events as far as I can tell.
LAMP however first picks AIMs (ancestry informmative markers) and goes trough the whole chromosome piece by piece in a moving window that changes in size depending on the physical position and the recombination rate of the window where it constantly compares to the given ancestral alleles for different populations. In this it differ from ADMIXTURE by looking at a group of SNP instead of a single SNP as ADMIXTURE. It comes without saying that comparing a group SNP gives a higher likelyhood of differentiating the actual origin of a segment than a single SNP.
This likely explain why some individuals that showed considerable admixture in ADMIXTURE had little or no such admixture in LAMP and why some proportions of other admixtures seen in ADMIXTURE was gone in LAMP. If looking only at single SNP's and averaging them some people may have f.ex "Saami" looking allele frequencies spread over the chromosome but when checked vs a segment of SNP's in the same segment they where actually more likely not of Saami origin.
In ADMIXTURE such problems as mention above go unnoticed unless the admixture is real and you can actually observe by comparing ADMIXTURE vs LAMP results that where the results appear consistent in both analysis its probably real or at least consistent given the parameters and data. This is because if the admixture is real a considerable of number of single SNP's with frequency close to a source populations will be there and it will of course by detected by ADMIXTURE.
Anders
The LAMP 2.5 genetic analysis software picks AIM's in its analysis based on the allele frequences given to it. On this basis I would like to check how LAMP perform by comparing to the widely used ADMIXTURE software. Of special interest are if ADMIXTURE clusters individuals similar to LAMP.
METHOD
I first estimatet the ancestral frequency data for Norwegians, Swedes, Saami and Finns from a subset of individuals clustering to these based on their MDS plots. I then run LAMP on recommended settings except this time I run it assuming the populations had been mixing for 10 generations the same presumption used in ADMIXTURE. I then picked the AIM's chosen for this analysis by LAMP and used them in a run for the whole dataset of 36 individuals.
OBSERVATIONS:
* All the Finns who clustered close to or 100% in LAMP also did the same in ADMIXTURE.
* In LAMP 9 Norwegians appeared 100% or close to 100% Norwegian. In ADMIXTURE 2 of these 9 appeared strongly mixed with Saami and Finns in ADMIXTURE.
* Saami all appear in LAMP close to or at 100% Saami. In ADMIXTURE only 1 appears as heavily mixed.
* All the Swedes who appeared as 100% Swedish in LAMP was also classified as 100% Swedish in ADMIXTURE.
* The Norwegian mixture in Swedes and Finns appears stronger in LAMP than in ADMIXTURE for those individuals who have it.
* Saami amix proportions seen in considerabe proportions in ADMIXTURE are often very little or gone in LAMP.
COMMENT:
It seems LAMP and ADMIXTURE is largely consistent about the "pure breed" and supports that the ancestral allele frequency data used in LAMP are correct. What LAMP and ADMIXTURE do disagree some about are the admixtures. 3 individuals who in ADMIXTURE was classified as mixed was not or very little admixted in LAMP. The reason for this are that LAMP and ADMIXTURE uses different methods to estimate admixture.
ADMIXTURE basically estimate allele freqency differences between SNP that is assumed to be independent and than cluster it. It does not care about physical positions of SNP, cM (centimorgans) between SNP and recombination events as far as I can tell.
LAMP however first picks AIMs (ancestry informmative markers) and goes trough the whole chromosome piece by piece in a moving window that changes in size depending on the physical position and the recombination rate of the window where it constantly compares to the given ancestral alleles for different populations. In this it differ from ADMIXTURE by looking at a group of SNP instead of a single SNP as ADMIXTURE. It comes without saying that comparing a group SNP gives a higher likelyhood of differentiating the actual origin of a segment than a single SNP.
This likely explain why some individuals that showed considerable admixture in ADMIXTURE had little or no such admixture in LAMP and why some proportions of other admixtures seen in ADMIXTURE was gone in LAMP. If looking only at single SNP's and averaging them some people may have f.ex "Saami" looking allele frequencies spread over the chromosome but when checked vs a segment of SNP's in the same segment they where actually more likely not of Saami origin.
In ADMIXTURE such problems as mention above go unnoticed unless the admixture is real and you can actually observe by comparing ADMIXTURE vs LAMP results that where the results appear consistent in both analysis its probably real or at least consistent given the parameters and data. This is because if the admixture is real a considerable of number of single SNP's with frequency close to a source populations will be there and it will of course by detected by ADMIXTURE.
Anders
mandag 18. april 2011
| I had earlier problems discriminating norwegians and swedes using the MDS plots and the ADMIXTURE program. This is have been problem that needed to be solved. I have for that reason (and some other reasons) learned a new genetic software called LAMP 2.5. This program have been specially designed to seperate individuals from closely related populations and it is able to specify what segment of the chromosome came from which population. The end result is quite similar to what is offered by 23andme's Ancestry Painting but instead of showing the dull labels "European", "African" and "East-Asian" you instead will see "Norwegian", "Swede", "Finn/Suomi" and "Saami" labels on your chromosome. In this test run I have limited the analysis to the X-chromosome but the analysis shouldnt be problem to escalate to the rest of the chromosome and to include more distant surrounding populations to investigate the relationship to continental Europe. The program may also be useful to detect specific segments for mixed dispora populations. The X-chromosome is a sex chromosome. It never pass two male generations. If you are a male you get your X-chromosome from your mother. If your female you get your fathers X-chromosome and one of the two of your mothers X-chromosome. If you are a male and have a Swedish mother and a Finnic father you would in a autosomal DNA analysis look like a 50/50 Swede/Finn but your X-chromosome you would appear as 100% Swedish because as a male you get your X-chromosome only from your mother. Whats do the analysis show? 1 The program is suprisingly good to label the main ancestry for each participants: a) All Norwegian participants where placed in correct category including two persons with large Saami and some Finnic mixture. b) Most Swedes participants where placed in correct category but 5 Swedes appeared to look more genetically similar to Norwegians than Swedes. This was also the case for a Swedish Finn (even a Estonian) who appeared to look more similar to Norwegians than to Swedes. Also worth to note that these Norwegian looking Swedes had some Finnic and even Saami admixture other Swedes didnt have. It may suggest that these participants have ancestry from the northern parts of Sweden. c) The Finns also have a good fit but there are some exceptions like the Swedish Finn mentioned above. This Finnic "outlieers" appears to come from border sones with other populations. d) The Saami appears to cluster nicely togheter. There appears to be some variation for one individual when assuming less time since the admixture event begun. There doesnt seem to have been more geneflow from scandinavians than finns or viceversa to the Saami. The most intriging question that arise from the above analysis are why do some swedes appears as more Norwegian than Swedish togheter with some Finnic and even Saami admixture in other words political borders do not seem to match the genetical borders completly. The traces of Finnic and Saami from this individuals suggest it is a northern phenomena in Sweden maybe from earlier migrations but it doesnt explain why a Swedish Finn and even a Estonian get so huge "Norwegian" percentages. Whats next? * More LAMP analyses trying to track origins to continental Europe * IBD analysis using newest technology. This will involve phasing of the genome for all participants. All will get their phased haplotypes for their chromosomes if they want. * ADMIXTURE analysis * MDS plots Anders |
torsdag 31. mars 2011
The 5th revision of the Fennoscandia Biographic Project
THIS IS THE LAST MAJOR REVISJON OF THE REGIONAL RESULT RELEASED 4TH DECEMBER 2010 DISTRIBUTED TO THE MEMBERS AT THAT TIME. IT DOES NOT CONTAIN ALL CURRENT MEMBERS.
Its finally time for a new update!
Whats new?
- Included reference populations from all over europe from Spain in the west to Cypriots in the south to Caucasus in the south-east and Chuvashes in the far east.
- ADMIXTURE run from K2 to K5
- MDS plots (earlier wrongly called PCA plots) up to 9 dimensions
- DIST plots similar to what i seen in 23andme. People typically match each other within Europe between 0.78 to 0.79. The differences is in these plots emphasised. There are 4 versions each sortert from the different nationalities Finns, Swedes, Norwegians and Saami.
Whats the result?
This is my suggested interpretation. For some of you these is probably much old news.
ADMIXTURE:
- At K2 or assuming two populations Europe is split into a southern (RED) and northern (BLUE) part with the Sardinians, Cypriots and the Caucasus populations cleary at the southern extreme while the Finns, Saami and the Chuvashes at the northern extreme. North in this context appears to point north-east geographically.
- At K3 or assuming three populations Europe is splitt into 1) south-east Europe populations (BLUE) the the Georgians at the extreme 2) south-west Europe populations (GREEN) with the Sardinians at the extreme 3) north/north-east Europe populations (RED) with the Saami at the extreme.
- At K4 or assuming four populations Europe is splitt into 1) Lithuanians at the extreme (BLUE) 2) Chuvasshes at the extreme (PINK) 3) Sardinians at the extreme (RED) 4) Georgians at the extreme (GREEN).
- At K5 or assuming four populations Europe splitt into 1) Basque at the extreme (RED) 2) Chuvashes at the extreme (PINK) 3) Lithuanians at the extreme (CYAN) 4) Sardinians at the extreme (BLUE) 5) Georgians at the extreme (GREEN).
COMMENT: At the highest K=5 the ADMIXTURE result *alone* seem to suggest that Norwegians, Swedes, Finna and Saami main ancestry is "Lithuanian" but that Finns and especially the Saami have a considerable "Chuvash" influences.
MDS-PLOTS:
- D1-2: The Norwegian and Swedish common clusters together with Belorussians and Lithuanians appear to bridge the space between Orcadians/French and Finns and Volgoda-Russians. The Saami appears to go further past the latter cluster outside towards the Chuvash. The Hungarians appear to neighbour the swedish/norwegian cluster to the upper-right.
- D1-3: The same story as above, but here the Chuvash share the far lower part of the plot with the Sardinians even the distances are large. Also here the Saami appear to pull from the Russians/Finns toward the Chuvashes. In the earlier plot the Chuvashes shared the higher part of the plot with Cacausus populations even the distances where large. The Lithuanians and Belorussians appear to seperate further lower away from the Finns and the Russians.
- D1-4: The same as above.
- D1-5: The Finns appears to have seperatet from the Russian and appears to pull toward the Saami who are alone at the extreme lower-left of the plot. Else much the same as above.
- D1-6: The Finns cluster with the Chuvashes with the Norwegian/Swedish cluster to the right. Russians and Lithuanians below. Saami alone at the upper-left extreme.
- D1-7: The Finns and the Saami appears to cluster alone to the lower-left. Norwegian-Swedish cluster partly with Belorussians/Orcadians.
- D1-8: Mostly the same as above.
- D1-9: Mostly the same as above.
COMMENT: The plots appears to show a more complex picture of the ancestry and relationship than the ADMIXTURE plots. In the lower dimensions the Saami appears to pull toward the Chuvashes but seem to seperate clearly often togheter alone with Finns in higher dimensions. Norwegians and Swedes appears mostly as expected to bridge between the more western populations with the more eastern.
DIST GRAPH:
The graph shows the average distance between populations seen from the four nationalities Norwegians, Swedish, Finns and Saami.
- Norwegians are more similar too Swedes, Belorussians, Orcadians and Lithuanians than themself (!). (Yes I have checked for errors).
- Swedes closest relatives are the Norwegians, Lithuanians and Belorussians. The most distant the Chuvashes, Cypriots and the Georgians.
- Finns closest relatives are the Lithuanians, Swedes and the Belorussians. The most distant are the Cypriots, Adigey and the Chuvashes.
- The Saamis clostest relatives are the Finns, Swedes and the Belorussians. The least distant the Cypriots, Armenians and the Georgians.
- There is very high correlation in the DIST plots between Norwegians and Swedes at 0.9842, then the Finns-Swedes at 0.9387, then Finns-Swedes at 0.9099, then Saami-Finns at 0.725, then Saami-Swedes at 0.511 and finally Saami-Norwegians at 0.4531.
- Saami appears to have consistently much lower genetic affinity with all other populations than the other three. The exception is to the Chuvashes where all appear to have somewhat similar genetic distance.
- Saami affiliation with continental and especially southern European populations are much smaller than the others.
Whats next?
- Get more samples
- Extend reference samples to include Euroasia
- Identify other analys methods
This concludes this round. There are no local MDS plot this time.
Anders
Its finally time for a new update!
Whats new?
- Included reference populations from all over europe from Spain in the west to Cypriots in the south to Caucasus in the south-east and Chuvashes in the far east.
- ADMIXTURE run from K2 to K5
- MDS plots (earlier wrongly called PCA plots) up to 9 dimensions
- DIST plots similar to what i seen in 23andme. People typically match each other within Europe between 0.78 to 0.79. The differences is in these plots emphasised. There are 4 versions each sortert from the different nationalities Finns, Swedes, Norwegians and Saami.
Whats the result?
This is my suggested interpretation. For some of you these is probably much old news.
ADMIXTURE:
- At K2 or assuming two populations Europe is split into a southern (RED) and northern (BLUE) part with the Sardinians, Cypriots and the Caucasus populations cleary at the southern extreme while the Finns, Saami and the Chuvashes at the northern extreme. North in this context appears to point north-east geographically.
- At K3 or assuming three populations Europe is splitt into 1) south-east Europe populations (BLUE) the the Georgians at the extreme 2) south-west Europe populations (GREEN) with the Sardinians at the extreme 3) north/north-east Europe populations (RED) with the Saami at the extreme.
- At K4 or assuming four populations Europe is splitt into 1) Lithuanians at the extreme (BLUE) 2) Chuvasshes at the extreme (PINK) 3) Sardinians at the extreme (RED) 4) Georgians at the extreme (GREEN).
- At K5 or assuming four populations Europe splitt into 1) Basque at the extreme (RED) 2) Chuvashes at the extreme (PINK) 3) Lithuanians at the extreme (CYAN) 4) Sardinians at the extreme (BLUE) 5) Georgians at the extreme (GREEN).
COMMENT: At the highest K=5 the ADMIXTURE result *alone* seem to suggest that Norwegians, Swedes, Finna and Saami main ancestry is "Lithuanian" but that Finns and especially the Saami have a considerable "Chuvash" influences.
MDS-PLOTS:
- D1-2: The Norwegian and Swedish common clusters together with Belorussians and Lithuanians appear to bridge the space between Orcadians/French and Finns and Volgoda-Russians. The Saami appears to go further past the latter cluster outside towards the Chuvash. The Hungarians appear to neighbour the swedish/norwegian cluster to the upper-right.
- D1-3: The same story as above, but here the Chuvash share the far lower part of the plot with the Sardinians even the distances are large. Also here the Saami appear to pull from the Russians/Finns toward the Chuvashes. In the earlier plot the Chuvashes shared the higher part of the plot with Cacausus populations even the distances where large. The Lithuanians and Belorussians appear to seperate further lower away from the Finns and the Russians.
- D1-4: The same as above.
- D1-5: The Finns appears to have seperatet from the Russian and appears to pull toward the Saami who are alone at the extreme lower-left of the plot. Else much the same as above.
- D1-6: The Finns cluster with the Chuvashes with the Norwegian/Swedish cluster to the right. Russians and Lithuanians below. Saami alone at the upper-left extreme.
- D1-7: The Finns and the Saami appears to cluster alone to the lower-left. Norwegian-Swedish cluster partly with Belorussians/Orcadians.
- D1-8: Mostly the same as above.
- D1-9: Mostly the same as above.
COMMENT: The plots appears to show a more complex picture of the ancestry and relationship than the ADMIXTURE plots. In the lower dimensions the Saami appears to pull toward the Chuvashes but seem to seperate clearly often togheter alone with Finns in higher dimensions. Norwegians and Swedes appears mostly as expected to bridge between the more western populations with the more eastern.
DIST GRAPH:
The graph shows the average distance between populations seen from the four nationalities Norwegians, Swedish, Finns and Saami.
- Norwegians are more similar too Swedes, Belorussians, Orcadians and Lithuanians than themself (!). (Yes I have checked for errors).
- Swedes closest relatives are the Norwegians, Lithuanians and Belorussians. The most distant the Chuvashes, Cypriots and the Georgians.
- Finns closest relatives are the Lithuanians, Swedes and the Belorussians. The most distant are the Cypriots, Adigey and the Chuvashes.
- The Saamis clostest relatives are the Finns, Swedes and the Belorussians. The least distant the Cypriots, Armenians and the Georgians.
- There is very high correlation in the DIST plots between Norwegians and Swedes at 0.9842, then the Finns-Swedes at 0.9387, then Finns-Swedes at 0.9099, then Saami-Finns at 0.725, then Saami-Swedes at 0.511 and finally Saami-Norwegians at 0.4531.
- Saami appears to have consistently much lower genetic affinity with all other populations than the other three. The exception is to the Chuvashes where all appear to have somewhat similar genetic distance.
- Saami affiliation with continental and especially southern European populations are much smaller than the others.
Whats next?
- Get more samples
- Extend reference samples to include Euroasia
- Identify other analys methods
This concludes this round. There are no local MDS plot this time.
Anders
The Fennoscandia Project blog
The Fennoscandia Biographic's Project's goal is the map the genetic relationship and origin of the current populations in Fennoscandia: The Norwegians, Swedes, Finns and the Saami.
The project was first announced on Rootsweb in 6th October 2010. The analysis and results have until now mainly been communicated trough Rootsweb-DNA postings and email correspondance. It currently has 41 members from the above mentioned countries and etnicites.
The analysis tools used so far have been genetic software like ADMIXTURE, PLINK's MDS plots and genetic similarity but other analytical tools will be applied at a later stage like IBD (Identity-By-Descent) estimation between individuals and origins on a chromosomal levels. Other analytical tools may be applied.
The project have a time limitation but is still open for participants of close to 100% Fennoscandian origin. Individuals of mixed Norwegian, Swedish, Finn or Saami origin are acceptable. Individuals of only partly Fennoscandian origins are not accepted in this project neiter is people of non-Fennoscandian origin.
If you fullfill the above requirement, have tested with 23andme, FamilyFinder or deCODEme and would like to participate please send your genome file in zipped form by email to tjaaehkere at yahoo no.
Please read the original Rootsweb-DNA posting for more details regarding the conditions for participating the project.
If any more questions or want to participate send email or email with your DNA file to:
tjaaehkere at yahoo.no (replace at with @)
This is the inital Rootsweb-DNA posting.
The project was first announced on Rootsweb in 6th October 2010. The analysis and results have until now mainly been communicated trough Rootsweb-DNA postings and email correspondance. It currently has 41 members from the above mentioned countries and etnicites.
The analysis tools used so far have been genetic software like ADMIXTURE, PLINK's MDS plots and genetic similarity but other analytical tools will be applied at a later stage like IBD (Identity-By-Descent) estimation between individuals and origins on a chromosomal levels. Other analytical tools may be applied.
The project have a time limitation but is still open for participants of close to 100% Fennoscandian origin. Individuals of mixed Norwegian, Swedish, Finn or Saami origin are acceptable. Individuals of only partly Fennoscandian origins are not accepted in this project neiter is people of non-Fennoscandian origin.
If you fullfill the above requirement, have tested with 23andme, FamilyFinder or deCODEme and would like to participate please send your genome file in zipped form by email to tjaaehkere at yahoo no.
Please read the original Rootsweb-DNA posting for more details regarding the conditions for participating the project.
If any more questions or want to participate send email or email with your DNA file to:
tjaaehkere at yahoo.no (replace at with @)
This is the inital Rootsweb-DNA posting.
Abonner på:
Kommentarer (Atom)